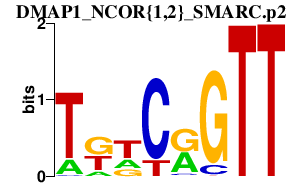
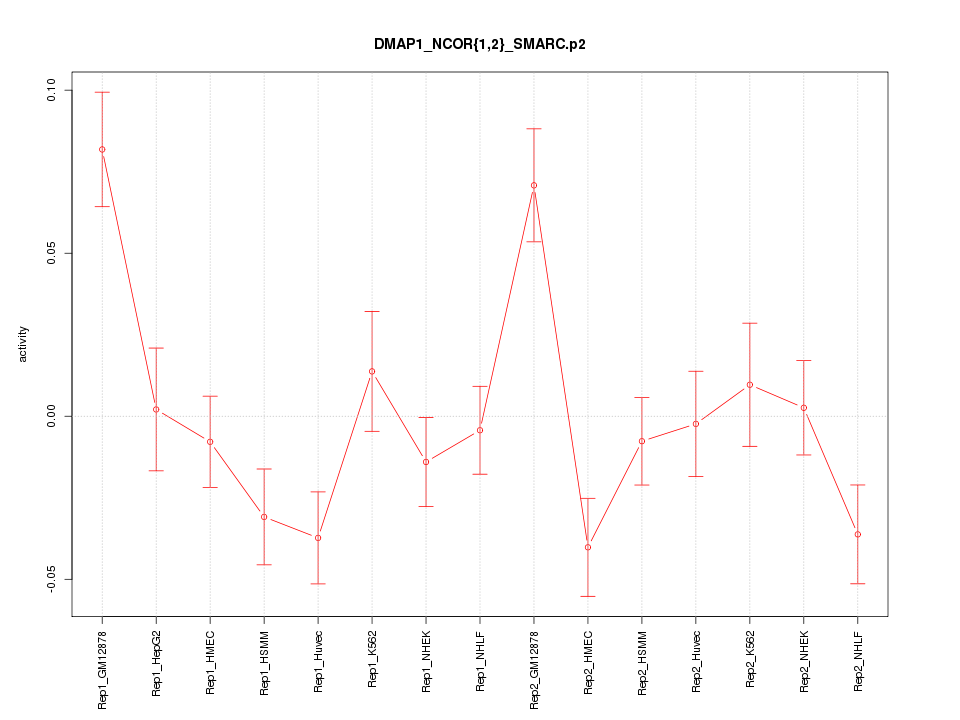
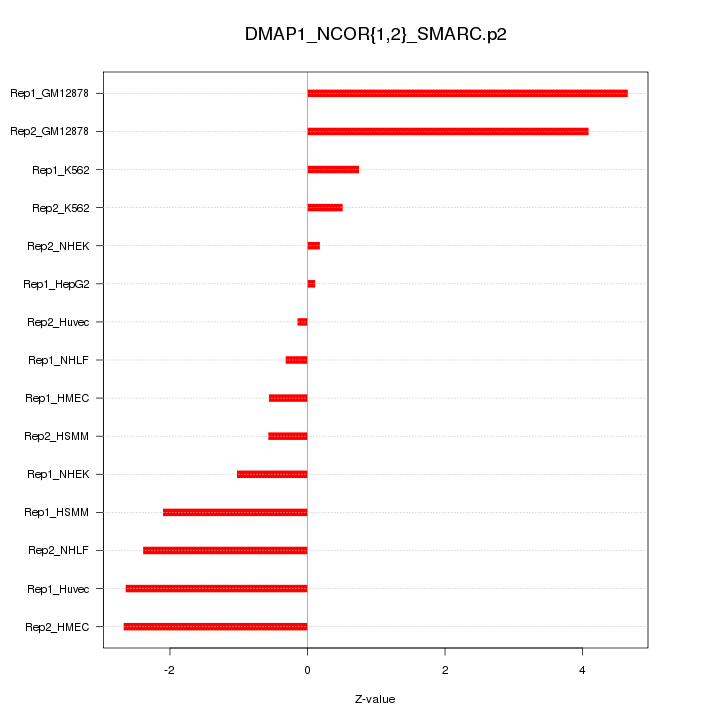
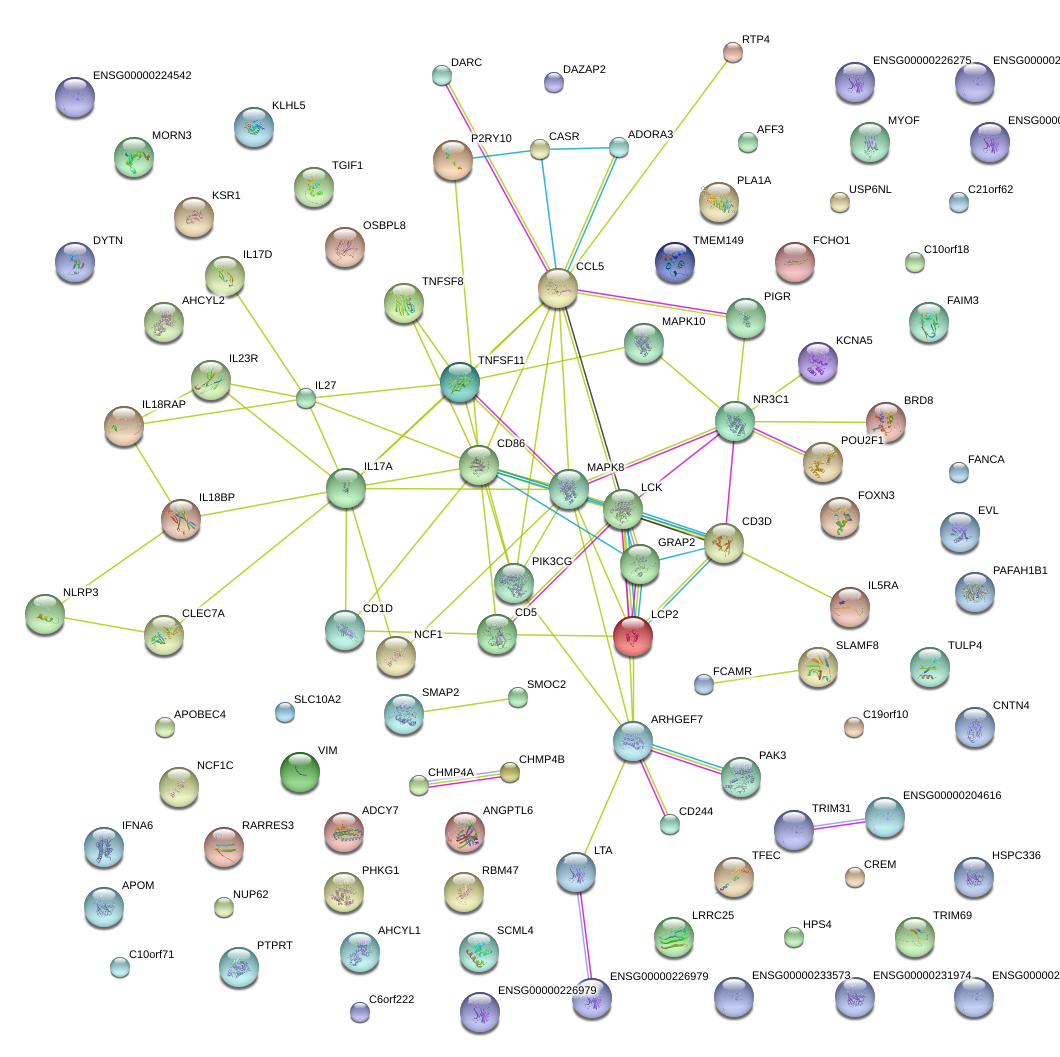
|
chr6_+_52159143
|
6.059
|
NM_002190
|
IL17A
|
interleukin 17A
|
|
chr15_+_42815851
|
5.317
|
NM_080745
NM_182985
|
TRIM69
|
tripartite motif containing 69
|
|
chr16_+_48865521
|
4.698
|
|
ADCY7
|
adenylate cyclase 7
|
|
chr3_+_120799411
|
4.510
|
NM_015900
|
PLA1A
|
phospholipase A1 member A
|
|
chr16_-_28425632
|
4.005
|
NM_145659
|
IL27
|
interleukin 27
|
|
chr7_+_73826244
|
3.825
|
NM_000265
|
NCF1
NCF1B
NCF1C
|
neutrophil cytosolic factor 1
neutrophil cytosolic factor 1B pseudogene
neutrophil cytosolic factor 1C pseudogene
|
|
chr1_+_165564904
|
3.630
|
NM_001198783
|
POU2F1
|
POU class 2 homeobox 1
|
|
chrX_+_78087484
|
3.505
|
NM_014499
NM_198333
|
P2RY10
|
purinergic receptor P2Y, G-protein coupled, 10
|
|
chr17_-_31231421
|
3.441
|
NM_002985
|
CCL5
|
chemokine (C-C motif) ligand 5
|
|
chr6_-_108252200
|
3.338
|
NM_198081
|
SCML4
|
sex comb on midleg-like 4 (Drosophila)
|
|
chr1_-_111848265
|
3.287
|
NM_000677
NM_020683
|
ADORA3
|
adenosine A3 receptor
|
|
chr1_+_245646080
|
3.246
|
NM_001079821
NM_001127461
|
NLRP3
|
NLR family, pyrin domain containing 3
|
|
chr19_-_55104875
|
3.195
|
|
NUP62
|
nucleoporin 62kDa
|
|
chr1_+_165565006
|
3.151
|
|
POU2F1
|
POU class 2 homeobox 1
|
|
chr6_-_36412562
|
3.032
|
NM_001010903
|
C6orf222
|
chromosome 6 open reading frame 222
|
|
chr17_+_22823151
|
3.020
|
NM_014238
|
KSR1
|
kinase suppressor of ras 1
|
|
chr22_+_38652555
|
3.006
|
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr5_-_138753443
|
2.984
|
NM_016459
|
MZB1
|
marginal zone B and B1 cell-specific protein
|
|
chr1_-_181889070
|
2.804
|
NM_203454
|
APOBEC4
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 4 (putative)
|
|
chr3_-_3127057
|
2.693
|
NM_000564
NM_175724
NM_175725
NM_175726
NM_175727
NM_175728
|
IL5RA
|
interleukin 5 receptor, alpha
|
|
chr5_-_169657395
|
2.676
|
NM_005565
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr5_-_169657312
|
2.671
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr7_+_106293112
|
2.635
|
NM_002649
|
PIK3CG
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr6_-_30188845
|
2.613
|
NM_007028
|
TRIM31
|
tripartite motif containing 31
|
|
chr1_+_156416360
|
2.577
|
NM_001766
|
CD1D
|
CD1d molecule
|
|
chr1_+_40635058
|
2.572
|
NM_001198980
|
SMAP2
|
small ArfGAP2
|
|
chr6_+_158653679
|
2.569
|
NM_001007466
NM_020245
|
TULP4
|
tubby like protein 4
|
|
chr10_-_11614279
|
2.515
|
NM_001080491
|
USP6NL
|
USP6 N-terminal like
|
|
chr2_+_102401580
|
2.473
|
NM_003853
|
IL18RAP
|
interleukin 18 receptor accessory protein
|
|
chr4_-_40211324
|
2.431
|
|
RBM47
|
RNA binding motif protein 47
|
|
chr19_-_10074332
|
2.298
|
NM_031917
|
ANGPTL6
|
angiopoietin-like 6
|
|
chr5_-_169657192
|
2.298
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr1_+_67404756
|
2.292
|
NM_144701
|
IL23R
|
interleukin 23 receptor
|
|
chr19_+_17723285
|
2.273
|
NM_001161359
NM_015122
|
FCHO1
|
FCH domain only 1
|
|
chr14_+_99601498
|
2.231
|
NM_016337
|
EVL
|
Enah/Vasp-like
|
|
chr1_+_32489426
|
2.218
|
NM_005356
|
LCK
|
lymphocyte-specific protein tyrosine kinase
|
|
chr16_-_88358946
|
2.191
|
|
FANCA
|
Fanconi anemia, complementation group A
|
|
chr6_+_31647854
|
2.175
|
NM_001159740
|
LTA
|
lymphotoxin alpha (TNF superfamily, member 1)
|
|
chr14_+_99601517
|
2.171
|
|
EVL
|
Enah/Vasp-like
|
|
chr13_-_102517196
|
2.166
|
NM_000452
|
SLC10A2
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 2
|
|
chr5_-_169657360
|
2.162
|
|
LCP2
|
lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa)
|
|
chr21_-_33107813
|
2.155
|
NM_001162495
NM_001162496
NM_019596
|
C21orf62
|
chromosome 21 open reading frame 62
|
|
chr1_-_205161821
|
2.144
|
NM_001193338
NM_001142473
NM_005449
|
FAIM3
|
Fas apoptotic inhibitory molecule 3
|
|
chr11_+_71387572
|
2.125
|
NM_001039659
NM_001039660
NM_173042
|
IL18BP
|
interleukin 18 binding protein
|
|
chr10_+_35466715
|
2.123
|
NM_183011
NM_183012
|
CREM
|
cAMP responsive element modulator
|
|
chr9_-_116732403
|
2.099
|
NM_001244
|
TNFSF8
|
tumor necrosis factor (ligand) superfamily, member 8
|
|
chr10_+_5812616
|
2.091
|
|
C10orf18
|
chromosome 10 open reading frame 18
|
|
chr3_+_2528272
|
2.089
|
|
CNTN4
|
contactin 4
|
|
chr13_+_42034871
|
2.083
|
NM_033012
|
TNFSF11
|
tumor necrosis factor (ligand) superfamily, member 11
|
|
chr22_-_25205376
|
2.030
|
NM_152841
|
HPS4
|
Hermansky-Pudlak syndrome 4
|
|
chr12_-_10173947
|
2.022
|
NM_022570
NM_197947
NM_197948
NM_197949
NM_197950
NM_197954
|
CLEC7A
|
C-type lectin domain family 7, member A
|
|
chr1_+_157441824
|
1.997
|
NM_001122951
|
DARC
|
Duffy blood group, chemokine receptor
|
|
chr1_-_205186330
|
1.975
|
NM_002644
|
PIGR
|
polymeric immunoglobulin receptor
|
|
chr10_-_95231933
|
1.949
|
NM_013451
NM_133337
|
MYOF
|
myoferlin
|
|
chr17_-_1195096
|
1.897
|
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa)
|
|
chr10_+_17312654
|
1.890
|
|
VIM
|
vimentin
|
|
chr6_+_31731647
|
1.888
|
NM_019101
|
APOM
|
apolipoprotein M
|
|
chr1_+_32512298
|
1.875
|
NM_001042771
|
LCK
|
lymphocyte-specific protein tyrosine kinase
|
|
chr18_+_3441589
|
1.872
|
NM_170695
NM_173210
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr5_-_142795269
|
1.772
|
NM_001018074
NM_001018075
NM_001018077
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor)
|
|
chr1_-_159099090
|
1.673
|
NM_001166663
NM_001166664
NM_016382
|
CD244
|
CD244 molecule, natural killer cell receptor 2B4
|
|
chr19_-_40925151
|
1.664
|
NM_024660
|
TMEM149
|
transmembrane protein 149
|
|
chr6_+_31731667
|
1.649
|
|
APOM
|
apolipoprotein M
|
|
chr7_+_128795178
|
1.620
|
NM_001130723
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr1_-_205210567
|
1.619
|
NM_001122979
NM_001170631
NM_032029
|
FCAMR
|
Fc receptor, IgA, IgM, high affinity
|
|
chr3_+_123279439
|
1.618
|
NM_006889
|
CD86
|
CD86 molecule
|
|
chr1_+_157441133
|
1.618
|
NM_002036
|
DARC
|
Duffy blood group, chemokine receptor
|
|
chrX_+_110252960
|
1.598
|
NM_001128168
NM_001128172
NM_001128173
|
PAK3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr2_-_100125402
|
1.575
|
NM_002285
|
AFF3
|
AF4/FMR2 family, member 3
|
|
chr4_+_38740246
|
1.573
|
NM_015990
NM_199039
|
KLHL5
|
kelch-like 5 (Drosophila)
|
|
chr9_-_21340885
|
1.530
|
NM_021002
|
IFNA6
|
interferon, alpha 6
|
|
chr19_-_18369407
|
1.520
|
NM_145256
|
LRRC25
|
leucine rich repeat containing 25
|
|
chr12_+_49922592
|
1.470
|
|
DAZAP2
|
DAZ associated protein 2
|
|
chr22_+_38627031
|
1.466
|
NM_004810
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr2_-_207291364
|
1.448
|
NM_001093730
|
DYTN
|
dystrotelin
|
|
chr11_+_60626505
|
1.444
|
NM_014207
|
CD5
|
CD5 molecule
|
|
chr3_+_188568861
|
1.437
|
NM_022147
|
RTP4
|
receptor (chemosensory) transporter protein 4
|
|
chr11_+_63060848
|
1.431
|
NM_004585
|
RARRES3
|
retinoic acid receptor responder (tazarotene induced) 3
|
|
chr10_+_50177192
|
1.422
|
NM_001135196
NM_199459
|
C10orf71
|
chromosome 10 open reading frame 71
|
|
chr12_+_5023345
|
1.397
|
NM_002234
|
KCNA5
|
potassium voltage-gated channel, shaker-related subfamily, member 5
|
|
chr7_+_128795280
|
1.380
|
|
AHCYL2
|
adenosylhomocysteinase-like 2
|
|
chr14_-_88948486
|
1.356
|
|
FOXN3
|
forkhead box N3
|
|
chr9_+_126460511
|
1.352
|
|
|
|
|
chr1_+_158063130
|
1.324
|
|
SLAMF8
|
SLAM family member 8
|
|
chr7_-_115458047
|
1.296
|
|
TFEC
|
transcription factor EC
|
|
chr14_-_23752875
|
1.266
|
NM_014169
|
CHMP4A
|
chromatin modifying protein 4A
|
|
chr11_-_117718532
|
1.265
|
NM_000732
NM_001040651
|
CD3D
|
CD3d molecule, delta (CD3-TCR complex)
|
|
chr4_-_87500201
|
1.261
|
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr6_-_76259976
|
1.257
|
NM_015687
|
FILIP1
|
filamin A interacting protein 1
|
|
chr7_-_86433139
|
1.249
|
NM_152748
|
KIAA1324L
|
KIAA1324-like
|
|
chr12_-_9777126
|
1.236
|
NM_172004
|
CLECL1
|
C-type lectin-like 1
|
|
chr20_+_56919233
|
1.225
|
|
GNAS
|
GNAS complex locus
|
|
chr1_+_158063278
|
1.219
|
|
SLAMF8
|
SLAM family member 8
|
|
chrX_-_131401319
|
1.217
|
NM_018388
NM_133486
|
MBNL3
|
muscleblind-like 3 (Drosophila)
|
|
chr7_-_115458064
|
1.206
|
|
TFEC
|
transcription factor EC
|
|
chr9_-_139047112
|
1.203
|
NM_004479
|
FUT7
|
fucosyltransferase 7 (alpha (1,3) fucosyltransferase)
|
|
chrX_+_119379967
|
1.176
|
NM_001142447
NM_012069
|
ATP1B4
|
ATPase, Na+/K+ transporting, beta 4 polypeptide
|
|
chr1_-_165754325
|
1.173
|
NM_000734
NM_198053
|
CD247
|
CD247 molecule
|
|
chr2_+_143603368
|
1.170
|
NM_018460
|
ARHGAP15
|
Rho GTPase activating protein 15
|
|
chr1_+_196874839
|
1.167
|
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chrX_+_117410693
|
1.158
|
|
WDR44
|
WD repeat domain 44
|
|
chr2_+_169637255
|
1.136
|
NM_001142270
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr16_-_4778317
|
1.129
|
NM_001154458
NM_144605
|
SEPT12
|
septin 12
|
|
chr3_+_45902999
|
1.129
|
NM_006641
NM_031200
|
CCR9
|
chemokine (C-C motif) receptor 9
|
|
chr4_-_186970366
|
1.118
|
NM_003603
NM_001145670
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr14_-_106184988
|
1.117
|
|
IGHA1
IGHG1
|
immunoglobulin heavy constant alpha 1
immunoglobulin heavy constant gamma 1 (G1m marker)
|
|
chr7_-_115458010
|
1.114
|
NM_001018058
NM_012252
|
TFEC
|
transcription factor EC
|
|
chrX_-_30505804
|
1.111
|
NM_025159
|
CXorf21
|
chromosome X open reading frame 21
|
|
chr13_-_25687062
|
1.110
|
|
RNF6
|
ring finger protein (C3H2C3 type) 6
|
|
chr3_+_46386636
|
1.100
|
NM_000579
NM_001100168
|
CCR5
|
chemokine (C-C motif) receptor 5
|
|
chr9_-_138762725
|
1.095
|
NM_198946
|
LCN6
|
lipocalin 6
|
|
chr2_+_143603439
|
1.090
|
|
ARHGAP15
|
Rho GTPase activating protein 15
|
|
chr2_+_228045053
|
1.084
|
|
AGFG1
|
ArfGAP with FG repeats 1
|
|
chr21_+_42697058
|
1.082
|
NM_001001895
NM_018961
|
UBASH3A
|
ubiquitin associated and SH3 domain containing A
|
|
chr16_+_10878539
|
1.071
|
NM_000246
|
CIITA
|
class II, major histocompatibility complex, transactivator
|
|
chr21_-_35181956
|
1.066
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr1_-_198643885
|
1.060
|
|
ZNF281
|
zinc finger protein 281
|
|
chr17_-_78000890
|
1.059
|
NM_001193655
|
C17orf62
|
chromosome 17 open reading frame 62
|
|
chr11_+_71388310
|
1.059
|
NM_001145057
|
IL18BP
|
interleukin 18 binding protein
|
|
chr7_+_77307382
|
1.054
|
NM_001127357
NM_001127359
NM_020432
|
PHTF2
|
putative homeodomain transcription factor 2
|
|
chr8_-_102872372
|
1.047
|
|
NCALD
|
neurocalcin delta
|
|
chr3_-_171070314
|
1.043
|
NM_024727
|
LRRC31
|
leucine rich repeat containing 31
|
|
chr1_-_246869181
|
1.041
|
NM_001001827
|
OR2T35
|
olfactory receptor, family 2, subfamily T, member 35
|
|
chr19_+_40895669
|
1.030
|
NM_014383
|
ZBTB32
|
zinc finger and BTB domain containing 32
|
|
chr3_-_114176401
|
1.023
|
NM_138806
NM_138939
NM_138940
NM_170780
|
CD200R1
|
CD200 receptor 1
|
|
chr21_+_42792810
|
1.022
|
NM_018964
|
SLC37A1
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 1
|
|
chr17_-_31441390
|
1.022
|
NM_002983
|
CCL3
|
chemokine (C-C motif) ligand 3
|
|
chr14_+_75058518
|
1.009
|
NM_006399
|
BATF
|
basic leucine zipper transcription factor, ATF-like
|
|
chr8_-_102872354
|
1.003
|
|
NCALD
|
neurocalcin delta
|
|
chr1_-_153098811
|
0.996
|
NM_170782
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3
|
|
chr2_-_74606819
|
0.994
|
NM_133637
|
DQX1
|
DEAQ box RNA-dependent ATPase 1
|
|
chr18_-_72820939
|
0.986
|
|
MBP
|
myelin basic protein
|
|
chr12_+_119562737
|
0.982
|
NM_001033677
|
CABP1
|
calcium binding protein 1
|
|
chr20_-_34707971
|
0.975
|
NM_032214
NM_175077
|
SLA2
|
Src-like-adaptor 2
|
|
chr14_-_106202262
|
0.975
|
|
IGLJ3
IGHA1
|
immunoglobulin lambda joining 3
immunoglobulin heavy constant alpha 1
|
|
chr12_+_91620749
|
0.964
|
NM_001037671
NM_001178097
|
C12orf74
|
chromosome 12 open reading frame 74
|
|
chr12_+_25096472
|
0.962
|
NM_006152
|
LRMP
|
lymphoid-restricted membrane protein
|
|
chr5_-_157011665
|
0.955
|
|
|
|
|
chr1_-_166149867
|
0.949
|
NM_001167749
NM_018417
|
ADCY10
|
adenylate cyclase 10 (soluble)
|
|
chr6_-_149847809
|
0.947
|
NM_207360
|
ZC3H12D
|
zinc finger CCCH-type containing 12D
|
|
chr4_-_164614335
|
0.939
|
NM_032136
|
TKTL2
|
transketolase-like 2
|
|
chr2_-_166359016
|
0.936
|
NM_004482
|
GALNT3
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3)
|
|
chr6_-_149847718
|
0.932
|
|
ZC3H12D
|
zinc finger CCCH-type containing 12D
|
|
chr2_-_74424041
|
0.924
|
NM_133478
|
SLC4A5
|
solute carrier family 4, sodium bicarbonate cotransporter, member 5
|
|
chr14_+_69307914
|
0.924
|
|
SRSF5
|
serine/arginine-rich splicing factor 5
|
|
chr7_+_28418668
|
0.916
|
NM_182898
|
CREB5
|
cAMP responsive element binding protein 5
|
|
chr12_-_8584658
|
0.911
|
NM_014358
|
CLEC4E
|
C-type lectin domain family 4, member E
|
|
chr11_+_118259684
|
0.905
|
NM_001716
|
CXCR5
|
chemokine (C-X-C motif) receptor 5
|
|
chr18_+_13601664
|
0.898
|
NM_001003674
NM_001003675
|
C18orf1
|
chromosome 18 open reading frame 1
|
|
chr22_+_38627129
|
0.896
|
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr16_+_24475129
|
0.896
|
|
RBBP6
|
retinoblastoma binding protein 6
|
|
chr1_+_173111306
|
0.892
|
NM_001035230
|
RABGAP1L
|
RAB GTPase activating protein 1-like
|
|
chr3_-_58175437
|
0.889
|
NM_004944
|
DNASE1L3
|
deoxyribonuclease I-like 3
|
|
chr1_+_154386358
|
0.886
|
NM_001193301
|
SEMA4A
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr3_+_112743615
|
0.873
|
NM_005816
NM_198196
|
CD96
|
CD96 molecule
|
|
chr6_+_99389318
|
0.863
|
|
POU3F2
|
POU class 3 homeobox 2
|
|
chr6_+_44076281
|
0.861
|
NM_001171992
NM_153246
|
C6orf223
|
chromosome 6 open reading frame 223
|
|
chr7_-_129379947
|
0.848
|
NM_003344
NM_182697
|
UBE2H
|
ubiquitin-conjugating enzyme E2H (UBC8 homolog, yeast)
|
|
chr1_+_196874759
|
0.844
|
NM_002838
NM_080921
NM_080923
|
PTPRC
|
protein tyrosine phosphatase, receptor type, C
|
|
chr2_+_218641982
|
0.841
|
NM_198483
|
RUFY4
|
RUN and FYVE domain containing 4
|
|
chr1_+_160868822
|
0.835
|
NM_001014796
NM_006182
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr22_-_16080265
|
0.825
|
|
CECR1
|
cat eye syndrome chromosome region, candidate 1
|
|
chrX_-_18282698
|
0.815
|
NM_006089
|
SCML2
|
sex comb on midleg-like 2 (Drosophila)
|
|
chr10_-_105208634
|
0.814
|
NM_001001412
|
CALHM1
|
calcium homeostasis modulator 1
|
|
chr14_-_105796268
|
0.814
|
|
ZCWPW2
IGHV3-23
IGKV3-20
|
zinc finger, CW type with PWWP domain 2
immunoglobulin heavy variable 3-23
immunoglobulin kappa variable 3-20
|
|
chr9_-_21218132
|
0.807
|
NM_021268
|
IFNA17
|
interferon, alpha 17
|
|
chr6_-_63053811
|
0.792
|
NM_152688
|
KHDRBS2
|
KH domain containing, RNA binding, signal transduction associated 2
|
|
chr12_-_119961143
|
0.791
|
NM_003733
NM_198213
|
OASL
|
2'-5'-oligoadenylate synthetase-like
|
|
chr3_+_179759181
|
0.787
|
NM_005832
|
KCNMB2
|
potassium large conductance calcium-activated channel, subfamily M, beta member 2
|
|
chr4_-_87500398
|
0.783
|
NM_138980
NM_138982
|
MAPK10
|
mitogen-activated protein kinase 10
|
|
chr1_-_210654858
|
0.778
|
|
TMEM206
|
transmembrane protein 206
|
|
chr1_+_163780063
|
0.774
|
NM_001005214
|
LRRC52
|
leucine rich repeat containing 52
|
|
chr2_-_215711313
|
0.772
|
NM_173076
|
ABCA12
|
ATP-binding cassette, sub-family A (ABC1), member 12
|
|
chr11_+_59001421
|
0.767
|
NM_001004705
|
OR4D10
|
olfactory receptor, family 4, subfamily D, member 10
|
|
chr1_+_156526182
|
0.766
|
NM_001765
|
CD1C
|
CD1c molecule
|
|
chr10_-_69125932
|
0.765
|
NM_013266
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3
|
|
chr1_-_210654862
|
0.759
|
NM_001198862
NM_018252
|
TMEM206
|
transmembrane protein 206
|
|
chr18_+_19706979
|
0.753
|
NM_000227
NM_001127718
|
LAMA3
|
laminin, alpha 3
|
|
chr4_+_147316284
|
0.743
|
NM_007080
|
LSM6
|
LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae)
|
|
chr1_+_160868932
|
0.739
|
|
DDR2
|
discoidin domain receptor tyrosine kinase 2
|
|
chr18_-_41923571
|
0.738
|
|
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle
|
|
chr9_+_37110506
|
0.737
|
|
ZCCHC7
|
zinc finger, CCHC domain containing 7
|
|
chr6_+_36773605
|
0.733
|
|
RAB44
|
RAB44, member RAS oncogene family
|
|
chr19_-_1046373
|
0.732
|
|
POLR2E
|
polymerase (RNA) II (DNA directed) polypeptide E, 25kDa
|
|
chr7_+_139750311
|
0.730
|
NM_001008749
|
RAB19
|
RAB19, member RAS oncogene family
|
|
chr16_-_49272741
|
0.727
|
NM_001144972
NM_153337
NM_182854
|
SNX20
|
sorting nexin 20
|
|
chr18_-_28606839
|
0.726
|
NM_020805
|
KLHL14
|
kelch-like 14 (Drosophila)
|
|
chr2_-_136311219
|
0.724
|
NM_002299
|
LCT
|
lactase
|
|
chr9_+_37110468
|
0.722
|
NM_032226
|
ZCCHC7
|
zinc finger, CCHC domain containing 7
|
|
chr2_+_68855436
|
0.720
|
NM_001166276
NM_014882
|
ARHGAP25
|
Rho GTPase activating protein 25
|
|
chr8_-_103735194
|
0.716
|
NM_001032282
|
KLF10
|
Kruppel-like factor 10
|
|
chr19_-_46504831
|
0.714
|
|
|
|
|
chr5_+_70787371
|
0.713
|
|
BDP1
|
B double prime 1, subunit of RNA polymerase III transcription initiation factor IIIB
|
|
chr8_-_38324809
|
0.709
|
|
WHSC1L1
|
Wolf-Hirschhorn syndrome candidate 1-like 1
|
|
chr2_-_152538694
|
0.707
|
NM_001005747
|
CACNB4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr21_-_31995972
|
0.701
|
|
SCAF4
|
SR-related CTD-associated factor 4
|
|
chr19_-_44518465
|
0.701
|
|
GMFG
|
glia maturation factor, gamma
|
|
chr17_+_55854636
|
0.696
|
NM_181707
|
C17orf64
|
chromosome 17 open reading frame 64
|
|
chr17_-_35327318
|
0.695
|
NM_018530
|
GSDMB
|
gasdermin B
|
|
chr7_+_94135384
|
0.694
|
|
PEG10
|
paternally expressed 10
|